Introduction
We suppose that you are already familiar with the Tutorial: Using Particle Filter to filter your data.
The Particle Filters (PF) are a set of Monte Carlo algorithms that permit to approximate solutions for filtering problems even when the state-space and/or measurement space are non-linear.
In this tutorial, we will use a PF to model a wire using polynomial interpolation . The PF is used to filter the noisy pixels in a segmented image in order to compute a model of the wire using polynomial interpolation. The color wire is observed by a static camera, in the following configuration:
The maths beyond the Particle Filter
The maths beyond the Particle Filter are explained in the documentation of the vpParticleFilter class. They are also explained in the The maths beyond the Particle Filter .
Explanations about the tutorial
The tutorial is split in three different programs:
How to run the tutorial
To run one of the tutorials with the default dataset, please run the following commands:
$ cd $VISP_WS/
visp-build/tutorial/particle-filter-curve-fitting
$ ./tutorial-pf-curve-fitting-{lms, pf, all} --video data/color_image_0%03d.png
To see the arguments the program can take, please run:
$ cd $VISP_WS/
visp-build/tutorial/particle-filter-curve-fitting
$ ./tutorial-pf-curve-fitting-{lms, pf, all} -h
You should see something similar to the following image:
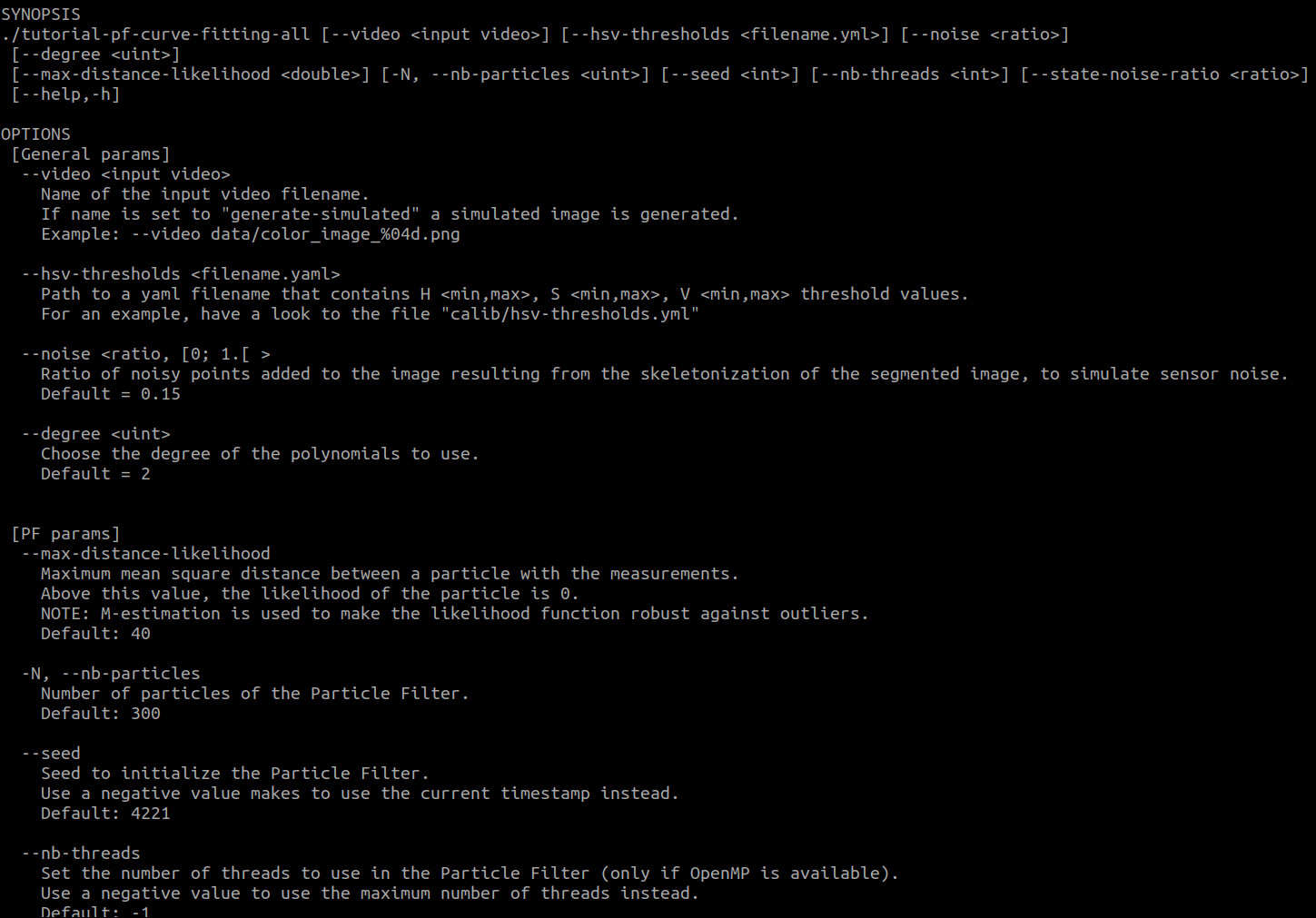
Screenshot of the tutorial Graphical User Interface
For the tutorial-pf-curve-fitting-pf.cpp and tutorial-pf-curve-fitting-all.cpp tutorials, the PF must be initialized. You are asked if you would rather use a manual initialization (left click) or an automatic one (right click).
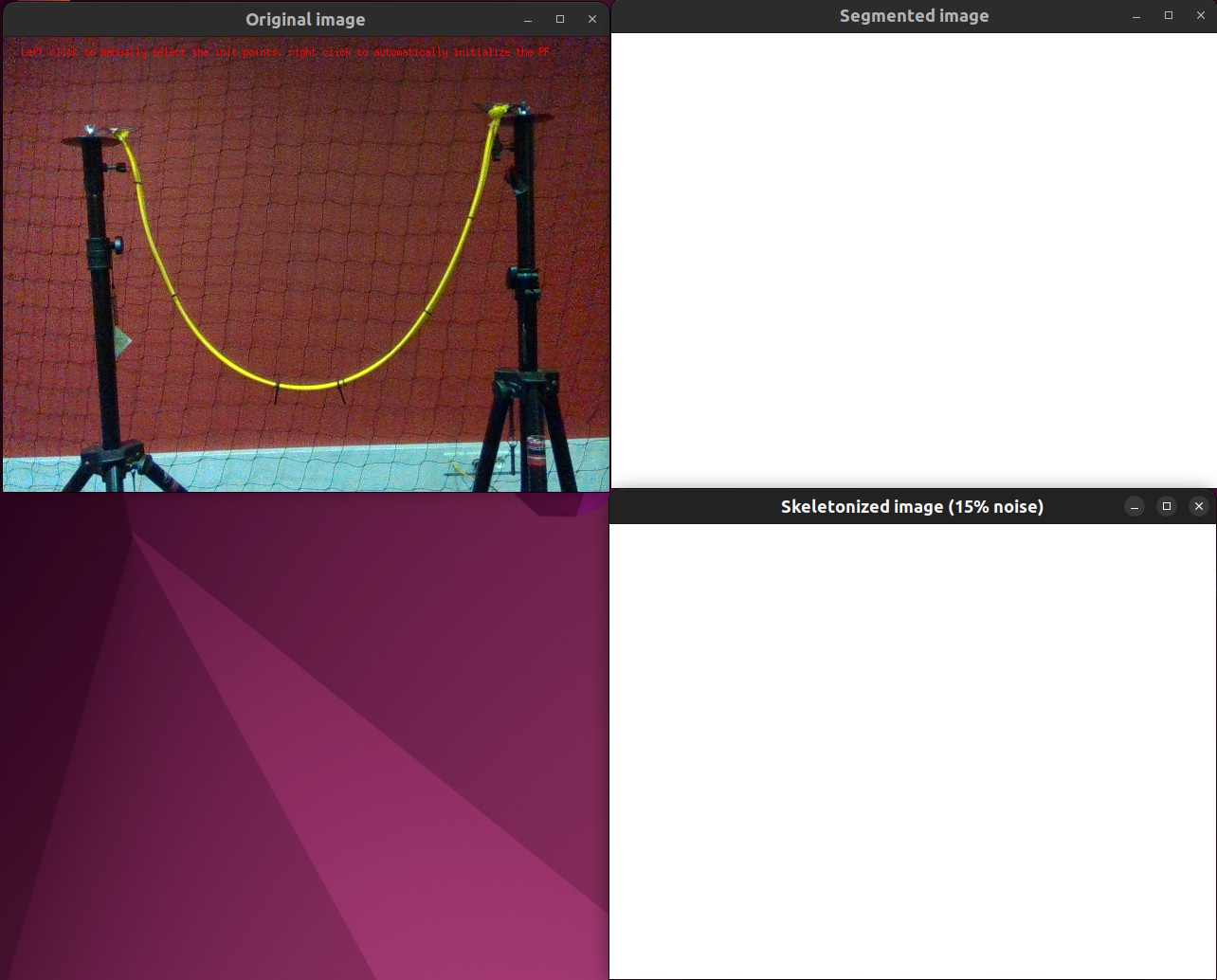
Screenshot of the tutorial initialization step
Then, for all the tutorials, you can either display the images one-by-one (left click) or automatically switch from an image to the next one (middle click). You can leave the program whenever you want using a right click.
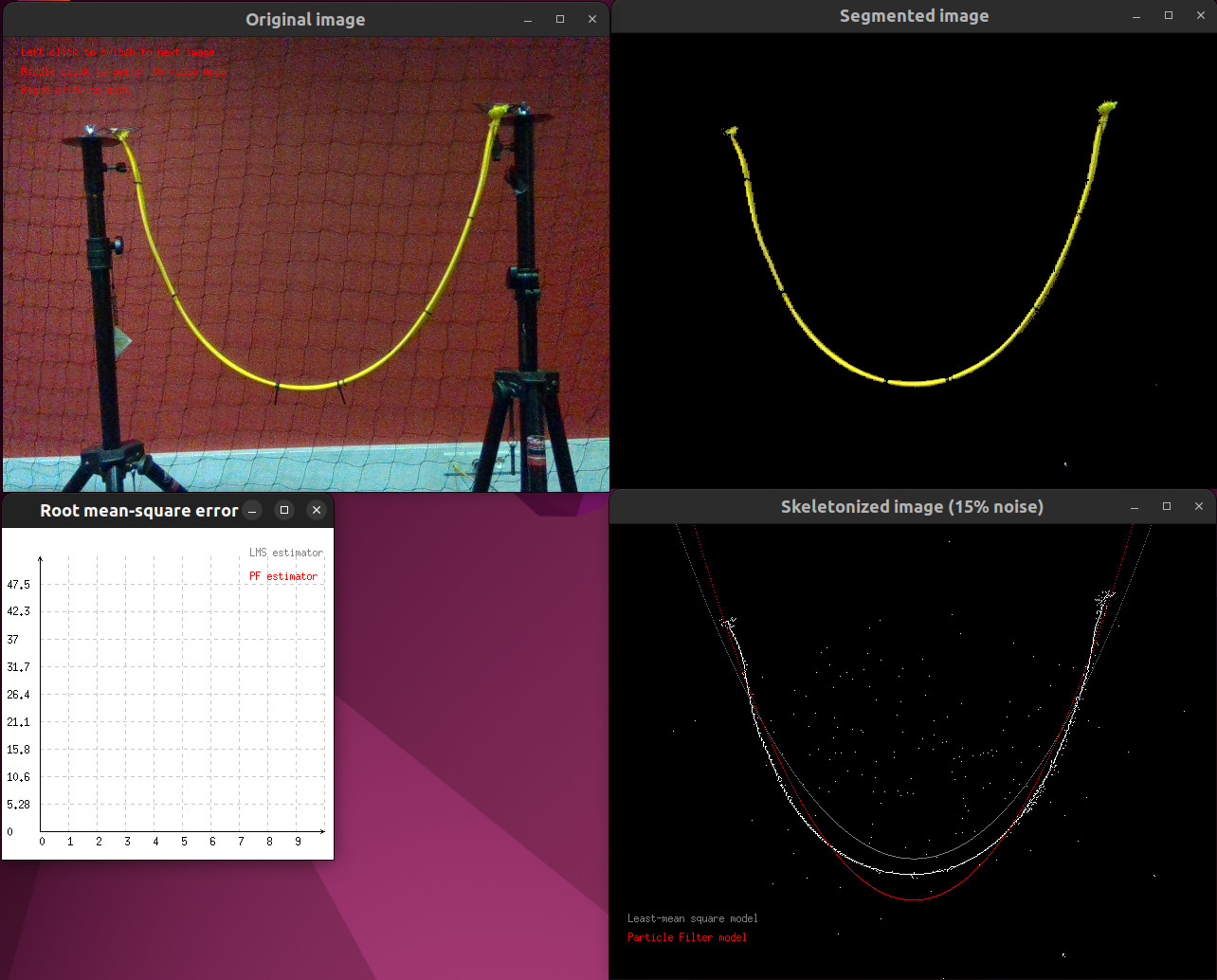
Screenshot of the tutorial during its run step
General explanations about the tutorials
Notes about the segmentation and skeletonization of the image
The segmentation of the image using HSV encoding has been presented in the Tutorial: HSV low/high range tuner tool .
A calibration file containing the segmentation parameters for the default sequence can be found in the calib folder. If you want to use your own sequence, please run the tutorial-hsv-range-tuner.cpp tutorial to extract the parameters that correspond to your own sequence and edit the file calib/hsv-thresholds.yml .
The skeletonization of the segmented image is not the topic of this tutorial. Thus, we invite the interested readers to read by themselves the corresponding method in the vpTutoSegmentation.cpp file.
Explanations about the Particle Filter
The internal state of the PF contains the coefficients of the interpolation polynomial. Be 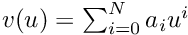 the interpolation polynomial whose highest degree is N. The internal state of the PF is of size N + 1 such as:
the interpolation polynomial whose highest degree is N. The internal state of the PF is of size N + 1 such as:
![\[ \begin{array}{lcl} \textbf{x}[i] &=& a_i \end{array} \]](form_1728.png)
The measurement 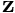 corresponds to the vector of vpImagePoint that forms the skeletonized version of the segmented image. Be
corresponds to the vector of vpImagePoint that forms the skeletonized version of the segmented image. Be 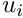 and
and 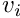 the horizontal and vertical pixel coordinates of the
the horizontal and vertical pixel coordinates of the 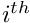 marker. The measurement vector can be written as:
marker. The measurement vector can be written as:
![\[ \begin{array}{lcl} \textbf{z}[i] &=& vpImagePoint(v_i , u_i) \end{array} \]](form_1729.png)
Explanations about the Least-Mean Square method
The Least-Mean Square method is a standard method to solve a polynomial interpolation problem. The polynomial interpolation problem can be written in matrices form as follow:
![\[ \begin{array}{llcl} & \textbf{A}\textbf{x} &=& \textbf{b} \\ \iff & \textbf{X} &=& \textbf{A}^+\textbf{b} \\ \end{array} \]](form_1730.png)
where 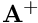 denotes the Moore-Penrose pseudo-inverse of the matrix A, with:
denotes the Moore-Penrose pseudo-inverse of the matrix A, with:
![\[ \begin{array}{lcl} \textbf{A}[i][j] &=& u_i^j \end{array} \\ \begin{array}{lcl} \textbf{x}[i] &=& a_i \end{array} \\ \begin{array}{lcl} \textbf{b}[i] &=& v_i \end{array} \]](form_1732.png)
Important note: due to numerical unstability, the pixel coordinates cannot be used directly in the Least-Mean square method. Indeed, using pixel coordinates leads to having an ill-conditionned  matrix. Consequently, in all the tutorials, the pixel coordinates are normalized by dividing them by the height and width of the image before running any interpolation.
matrix. Consequently, in all the tutorials, the pixel coordinates are normalized by dividing them by the height and width of the image before running any interpolation.
Detailed explanations about the tutorials
We will now present the different tutorials and explained their important parts.
Details on the tutorial-pf-curve-fitting-lms.cpp
This tutorial goal is to show how a Least-Mean Square method can be used to perform polynomial interpolation and the impact of noise on such technique.
To visualize the accuracy of the method, a vpPlot is instantiated in the following part of the code:
#ifdef VISP_HAVE_DISPLAY
unsigned int plotHeight = 350, plotWidth = 350;
int plotXpos = static_cast<int>(data.m_legendOffset.get_u());
int plotYpos = static_cast<int>(data.m_I_orig.getHeight() + 4. * data.m_legendOffset.get_v());
vpPlot plot(1, plotHeight, plotWidth, plotXpos, plotYpos,
"Root mean-square error");
plot.initGraph(0, 1);
plot.setLegend(0, 0, "LMS estimator");
#endif
static const vpColor gray
This class enables real time drawing of 2D or 3D graphics. An instance of the class open a window whi...
At each iteration, the new frame is read, then segmented and skeletonized in the following section of code:
std::cout << "Iter " << nbIter << std::endl;
data.m_grabber.acquire(data.m_I_orig);
tutorial::performSegmentationHSV(data);
std::vector<vpImagePoint> edgePoints = tutorial::extractSkeleton(data);
std::vector<vpImagePoint> noisyEdgePoints = tutorial::addSaltAndPepperNoise(edgePoints, data);
The method addSaltAndPepperNoise permits to add a user-defined percentage of salt-and-pepper noise, to evaluate the robustness of the method against noise.
The polynomial interpolation is performed in the following section of the code:
lmsFitter.fit(noisyEdgePoints);
It relies on the following method of the vpTutoMeanSquareFitting class:
void vpTutoMeanSquareFitting::fit(const std::vector<vpImagePoint> &pts)
{
vpTutoParabolaModel::fillSystem(m_degree, m_height, m_width, pts, A, b);
m_model = vpTutoParabolaModel(X, m_height, m_width);
m_isFitted = true;
}
Implementation of a matrix and operations on matrices.
vpMatrix pseudoInverse(double svThreshold=1e-6) const
The matrices that form the system that needs to be solved are filled in the vpTutoParabolaModel class (we remind the reader that normalization of the pixel coordinates is performed to avoid numerical unstability):
static void fillSystem(
const unsigned int °ree,
const double &height,
const double &width,
const std::vector<VISP_NAMESPACE_ADDRESSING vpImagePoint> &pts, VISP_NAMESPACE_ADDRESSING
vpMatrix &A, VISP_NAMESPACE_ADDRESSING
vpMatrix &b)
{
const unsigned int nbPts = static_cast<unsigned int>(pts.size());
const unsigned int nbCoeffs = degree + 1;
std::vector<VISP_NAMESPACE_ADDRESSING vpImagePoint> normalizedPts;
for (const auto &pt: pts) {
normalizedPts.push_back(VISP_NAMESPACE_ADDRESSING
vpImagePoint(pt.get_i() / height, pt.get_j() / width));
}
A.resize(nbPts, nbCoeffs, 1.);
b.resize(nbPts, 1);
for (unsigned int i = 0; i < nbPts; ++i) {
double u = normalizedPts[i].get_u();
double v = normalizedPts[i].get_v();
for (unsigned int j = 0; j < nbCoeffs; ++j) {
A[i][j] = std::pow(u, j);
}
b[i][0] = v;
}
}
Class that defines a 2D point in an image. This class is useful for image processing and stores only ...
Details on the tutorial-pf-curve-fitting-pf.cpp
This tutorial is meant to show how a Particle Filter could be used to model a wire thanks to polynomial interpolation.
To initialize a Particle Filter, a guess of the initial state must be given to it. The more accurate is the initial guess, the quicker the PF will converge. The initialization can be done either manually or automatically thanks to the following piece of code:
std::vector<vpImagePoint> automaticInitialization(tutorial::vpTutoCommonData &data)
{
const unsigned int minNbPts = data.m_degree + 1;
const unsigned int nbPtsToUse = 10 * minNbPts;
std::vector<vpImagePoint> initPoints;
tutorial::performSegmentationHSV(data);
std::vector<vpImagePoint> edgePoints = tutorial::extractSkeleton(data);
unsigned int nbEdgePoints = static_cast<unsigned int>(edgePoints.size());
if (nbEdgePoints < nbPtsToUse) {
return edgePoints;
}
return ptA.
get_u() < ptB.get_u();
};
std::sort(edgePoints.begin(), edgePoints.end(), ptHasLowerU);
unsigned int idStart, idStop;
if (nbEdgePoints > nbPtsToUse + 20) {
idStart = 10;
idStop = static_cast<unsigned int>(edgePoints.size()) - 10;
}
else {
idStart = 0;
idStop = static_cast<unsigned int>(edgePoints.size());
}
unsigned int sizeWindow = idStop - idStart + 1;
unsigned int step = sizeWindow / (nbPtsToUse - 1);
for (unsigned int id = idStart; id <= idStop; id += step) {
initPoints.push_back(edgePoints[id]);
}
return initPoints;
}
std::vector<vpImagePoint> manualInitialization(const tutorial::vpTutoCommonData &data)
{
const bool waitForClick = true;
const unsigned int sizeCross = 10;
const unsigned int thicknessCross = 2;
const unsigned int minNbPts = data.m_degree + 1;
std::vector<vpImagePoint> initPoints;
bool notEnoughPoints = true;
while (notEnoughPoints) {
vpDisplay::displayText(data.m_I_orig, data.m_ipLegend,
"Left click to add init point (min.: " + std::to_string(minNbPts) +
"), right click to estimate the initial coefficients of the Particle Filter.", data.m_colorLegend);
vpDisplay::displayText(data.m_I_orig, data.m_ipLegend + data.m_legendOffset,
"A middle click reinitialize the list of init points.", data.m_colorLegend);
vpDisplay::displayText(data.m_I_orig, data.m_ipLegend + data.m_legendOffset + data.m_legendOffset,
"If not enough points have been selected, a right click has no effect.", data.m_colorLegend);
unsigned int nbInitPoints = static_cast<unsigned int>(initPoints.size());
for (unsigned int i = 0; i < nbInitPoints; ++i) {
}
switch (button) {
case vpMouseButton::vpMouseButtonType::button1:
initPoints.push_back(ipClick);
break;
case vpMouseButton::vpMouseButtonType::button2:
initPoints.clear();
break;
case vpMouseButton::vpMouseButtonType::button3:
(initPoints.size() >= minNbPts ? notEnoughPoints = false : notEnoughPoints = true);
break;
default:
break;
}
}
return initPoints;
}
vpColVector computeInitialGuess(tutorial::vpTutoCommonData &data)
{
std::vector<vpImagePoint> initPoints;
#ifdef VISP_HAVE_DISPLAY
const bool waitForClick = true;
const unsigned int sizeCross = 10;
const unsigned int thicknessCross = 2;
bool automaticInit = false;
vpDisplay::displayText(data.m_I_orig, data.m_ipLegend,
"Left click to manually select the init points, right click to automatically initialize the PF", data.m_colorLegend);
switch (button) {
case vpMouseButton::vpMouseButtonType::button1:
automaticInit = false;
break;
case vpMouseButton::vpMouseButtonType::button3:
automaticInit = true;
break;
default:
break;
}
if (automaticInit) {
initPoints = tutorial::automaticInitialization(data);
}
else {
initPoints = tutorial::manualInitialization(data);
}
#else
initPoints = tutorial::automaticInitialization(data);
#endif
tutorial::vpTutoMeanSquareFitting lmsFitter(data.m_degree, data.m_I_orig.getHeight(), data.m_I_orig.getWidth());
lmsFitter.fit(initPoints);
std::cout << "---[Initial fit]---" << std::endl;
std::cout << lmsFitter.getModel();
std::cout << "---[Initial fit]---" << std::endl;
vpDisplay::displayText(data.m_I_orig, data.m_ipLegend,
"Here are the points selected for the initialization.", data.m_colorLegend);
unsigned int nbInitPoints = static_cast<unsigned int>(initPoints.size());
for (unsigned int i = 0; i < nbInitPoints; ++i) {
}
lmsFitter.display(data.m_I_orig,
vpColor::red,
static_cast<unsigned int>(data.m_ipLegend.get_v() + 2 * data.m_legendOffset.get_v()),
static_cast<unsigned int>(data.m_ipLegend.get_u()));
vpDisplay::displayText(data.m_I_orig, data.m_ipLegend + data.m_legendOffset,
"A click to continue.", data.m_colorLegend);
return X0;
}
Implementation of column vector and the associated operations.
Class to define RGB colors available for display functionalities.
static bool getClick(const vpImage< unsigned char > &I, bool blocking=true)
static void display(const vpImage< unsigned char > &I)
static void displayCross(const vpImage< unsigned char > &I, const vpImagePoint &ip, unsigned int size, const vpColor &color, unsigned int thickness=1)
static void flush(const vpImage< unsigned char > &I)
static void displayText(const vpImage< unsigned char > &I, const vpImagePoint &ip, const std::string &s, const vpColor &color)
A Particle Filter needs a function to evaluate the likelihood of a particle. We decided to use a Gaussian-based function that penalizes particles whose polynomial model has a Root Mean Square Error with regard to the measurement points greater than a given threshold. To be robust against outliers, we use the Tukey M-estimator. The likelihood function is implemented in a functor to be able to pass additional information such as the height and width of the input image:
class vpTutoLikelihoodFunctor
{
public:
vpTutoLikelihoodFunctor(const double &stdev, const unsigned int &height, const unsigned int &width)
: m_height(height)
, m_width(width)
{
double sigmaDistanceSquared = stdev * stdev;
m_constantDenominator = 1. / std::sqrt(2. * M_PI * sigmaDistanceSquared);
m_constantExpDenominator = -1. / (2. * sigmaDistanceSquared);
}
double likelihood(
const vpColVector &coeffs,
const std::vector<vpImagePoint> &meas)
{
double likelihood = 0.;
unsigned int nbPoints = static_cast<unsigned int>(meas.size());
vpTutoParabolaModel model(coeffs, m_height, m_width);
for (unsigned int i = 0; i < nbPoints; ++i) {
double squareError = tutorial::evaluate(meas[i], model);
residuals[i] = squareError;
}
double sumError = w.hadamard(residuals).sum();
likelihood = std::exp(m_constantExpDenominator * sumError / w.sum()) * m_constantDenominator;
likelihood = std::min(likelihood, 1.0);
likelihood = std::max(likelihood, 0.);
return likelihood;
}
private:
double m_constantDenominator;
double m_constantExpDenominator;
unsigned int m_height;
unsigned int m_width;
};
Contains an M-estimator and various influence function.
@ TUKEY
Tukey influence function.
void MEstimator(const vpRobustEstimatorType method, const vpColVector &residues, vpColVector &weights)
This likelihood functor compute the residuals using the following evaluation functions:
double evaluate(
const vpImagePoint &pt,
const vpTutoParabolaModel &model)
{
double v_model = model.eval(u);
double error = v - v_model;
double squareError = error * error;
return squareError;
}
double evaluate(
const vpColVector &coeffs,
const unsigned int &height,
const unsigned int &width,
const std::vector<vpImagePoint> &pts)
{
unsigned int nbPts = static_cast<unsigned int>(pts.size());
vpTutoParabolaModel model(coeffs, height, width);
for (unsigned int i = 0; i < nbPts; ++i) {
double squareError = evaluate(pts[i], model);
residuals[i] = squareError;
}
double meanSquareError = residuals.sum() / static_cast<double>(nbPts);
return std::sqrt(meanSquareError);
}
A Particle Filter needs to perform a weighted average to compute the filtered state. Performing a weighted average of the particles polynomial coefficients would not lead to satisfying results, as it is not mathematically correct. Instead, we decided to generate a given number of "control points" by each particle. The number of control points generated by a particle is dictated by its associated weight. Then, we compute the polynomial coefficients that best fit all these control points using a Least-Square minimization technique. The weighted average is performed thanks to the following functor:
class vpTutoAverageFunctor
{
public:
vpTutoAverageFunctor(const unsigned int °ree, const unsigned int &height, const unsigned int &width)
: m_degree(degree)
, m_height(height)
, m_width(width)
{ }
vpColVector averagePolynomials(
const std::vector<vpColVector> &particles,
const std::vector<double> &weights,
const vpParticleFilter<std::vector<vpImagePoint>>::vpStateAddFunction &)
{
const unsigned int nbParticles = static_cast<unsigned int>(particles.size());
const double nbParticlesAsDOuble = static_cast<double>(nbParticles);
const double sumWeight = std::accumulate(weights.begin(), weights.end(), 0.);
const double nbPointsForAverage = 10. * nbParticlesAsDOuble;
std::vector<vpImagePoint> initPoints;
for (unsigned int i = 0; i < nbParticles; ++i) {
double nbPoints = std::floor(weights[i] * nbPointsForAverage / sumWeight);
if (nbPoints > 1.) {
vpTutoParabolaModel curve(particles[i], m_height, m_width);
double widthAsDouble = static_cast<double>(m_width);
double step = widthAsDouble / (nbPoints - 1.);
for (double u = 0.; u < widthAsDouble; u += step) {
double v = curve.eval(u);
initPoints.push_back(pt);
}
}
else if (nbPoints == 1.) {
vpTutoParabolaModel curve(particles[i], m_height, m_width);
double u = static_cast<double>(m_width) / 2.;
double v = curve.eval(u);
initPoints.push_back(pt);
}
}
vpTutoMeanSquareFitting lms(m_degree, m_height, m_width);
lms.fit(initPoints);
return lms.getCoeffs();
}
private:
unsigned int m_degree;
unsigned int m_height;
unsigned int m_width;
};
The class permits to use a Particle Filter.
A Particle Filter needs a process function to project the particles forward in time. For this scenario, we decided to use the identity, and let the randomness of the Particle Filter manage the potential motion of the wire.
{
return updatedCoeffs;
}
The initialization parameters of the Particle Filter are defined in the following section of code:
const double maxDistanceForLikelihood = data.m_pfMaxDistanceForLikelihood;
const double sigmaLikelihood = maxDistanceForLikelihood / 3.;
const unsigned int nbParticles = data.m_pfN;
std::vector<double> stdevsPF;
for (unsigned int i = 0; i < data.m_degree + 1; ++i) {
double ampliMax = data.m_pfRatiosAmpliMax[i] * X0[i];
stdevsPF.push_back(ampliMax / 3.);
}
unsigned long seedPF;
const float period = 33.3f;
if (data.m_pfSeed < 0) {
}
else {
seedPF = data.m_pfSeed;
}
const int nbThread = data.m_pfNbThreads;
VISP_EXPORT double measureTimeMicros()
The initialization functions of the Particle Filter are defined in the following section of code:
tutorial::vpTutoLikelihoodFunctor likelihoodFtor(sigmaLikelihood, data.m_I_orig.getHeight(), data.m_I_orig.getWidth());
using std::placeholders::_1;
using std::placeholders::_2;
tutorial::vpTutoAverageFunctor averageCpter(data.m_degree, data.m_I_orig.getHeight(), data.m_I_orig.getWidth());
using std::placeholders::_3;
The Particle Filter is then constructed thanks to the following lines:
filter.init(X0, processFunc, likelihoodFunc, checkResamplingFunc, resamplingFunc, meanFunc);
Finally, the filtering is performed thanks to the following line:
filter.filter(noisyEdgePoints, period);
The filtered state is retrieve thanks to the following line:
Details on the tutorial-pf-curve-fitting-all.cpp
The tutorial-pf-curve-fitting-all.cpp reuse what has already been presented in the Details on the tutorial-pf-curve-fitting-lms.cpp and Details on the tutorial-pf-curve-fitting-pf.cpp sections.
Its main objective is to compare the time performances and robustness against noise of the two methods. The ratio of noise can be set using the Command Line Interface using the –noise option. For instance,
$ ./tutorial-pf-curve-fitting-all --video data/color_image_0%03d.png --noise <ratio in the intervall [0.; 1.[>
will add 50% of noisy pixels to the skeletonized image.
You can experiment by varying this parameter (and others as well) to see the impact on the different methods. With 15% noisy pixels and more, the Particle Filter tends to have a greater accuracy than the LMS method, but it takes more times to run.






![]() the interpolation polynomial whose highest degree is N. The internal state of the PF is of size N + 1 such as:
the interpolation polynomial whose highest degree is N. The internal state of the PF is of size N + 1 such as:![]()
![]() corresponds to the vector of vpImagePoint that forms the skeletonized version of the segmented image. Be
corresponds to the vector of vpImagePoint that forms the skeletonized version of the segmented image. Be ![]() and
and ![]() the horizontal and vertical pixel coordinates of the
the horizontal and vertical pixel coordinates of the ![]() marker. The measurement vector can be written as:
marker. The measurement vector can be written as:![]()
![]()
![]() denotes the Moore-Penrose pseudo-inverse of the matrix A, with:
denotes the Moore-Penrose pseudo-inverse of the matrix A, with:![]()
![]() matrix. Consequently, in all the tutorials, the pixel coordinates are normalized by dividing them by the height and width of the image before running any interpolation.
matrix. Consequently, in all the tutorials, the pixel coordinates are normalized by dividing them by the height and width of the image before running any interpolation.